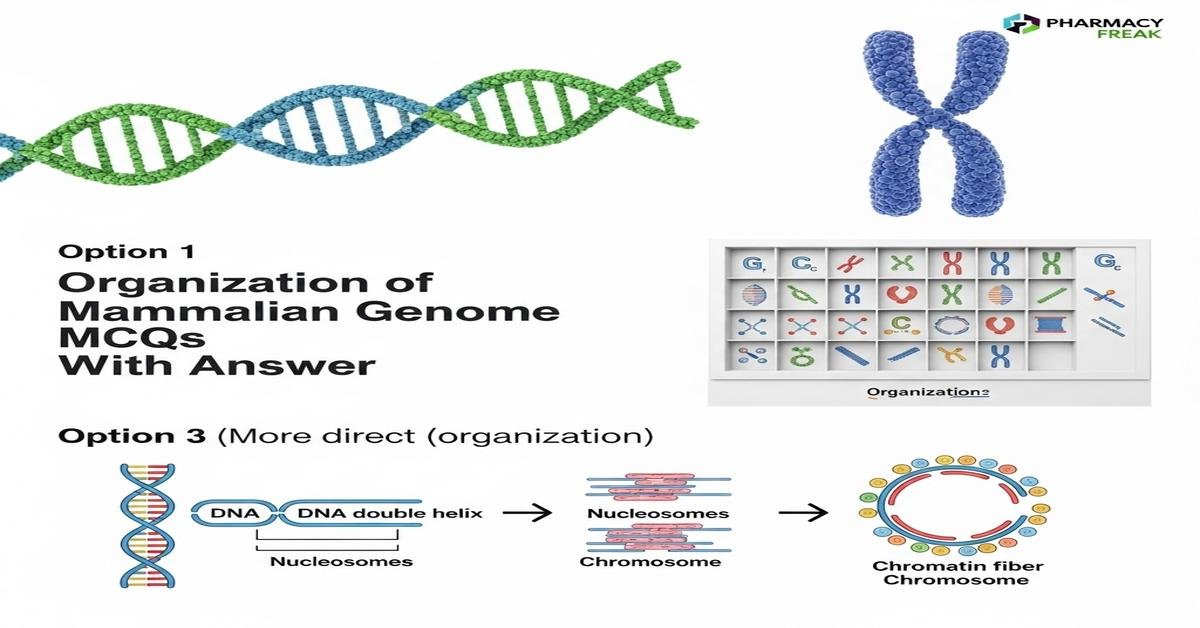
Organization of mammalian genome MCQs With Answer is a focused learning resource for B. Pharm students that explains genome structure, chromatin architecture, gene regulation, and epigenetic controls in mammals. This Student-friendly post highlights key concepts like nucleosomes, histone modifications, heterochromatin vs euchromatin, regulatory elements (promoters, enhancers, insulators), telomeres, centromeres, and 3D genome organization (TADs, loops). These topics are essential for understanding pharmacogenomics, drug targets, and gene expression relevant to pharmacy practice. The material includes multiple-choice questions with answers to reinforce core principles and prepare students for exams. Now let’s test your knowledge with 50 MCQs on this topic.
Q1. Which structure is the basic repeating unit of chromatin in mammalian cells?
- Nucleosome
- Sarcomere
- Ribosome
- Centromere
Correct Answer: Nucleosome
Q2. What histone proteins form the core around which DNA is wrapped in a nucleosome?
- H1 only
- H2A, H2B, H3, H4
- H5 and protamines
- H2A and H2B only
Correct Answer: H2A, H2B, H3, H4
Q3. Which histone is commonly associated with linker DNA and helps compact chromatin?
- H3
- H1
- H2B
- H4
Correct Answer: H1
Q4. DNA methylation in mammals most often occurs at which sequence context?
- GpC sites
- CpG sites
- ApT sites
- TpA sites
Correct Answer: CpG sites
Q5. Which chromatin state is typically transcriptionally active?
- Heterochromatin
- Centromeric chromatin
- Euchromatin
- Satellite DNA
Correct Answer: Euchromatin
Q6. Which enzyme adds acetyl groups to histone tails, generally activating transcription?
- DNA methyltransferase (DNMT)
- Histone deacetylase (HDAC)
- Histone acetyltransferase (HAT)
- Topoisomerase
Correct Answer: Histone acetyltransferase (HAT)
Q7. What is the functional role of CpG islands in mammalian promoters?
- Serve as replication origins only
- Are binding sites for ribosomes
- Often mark promoters and regulate transcription via methylation
- Encode microRNAs
Correct Answer: Often mark promoters and regulate transcription via methylation
Q8. Which protein is a well-known insulator binding factor involved in 3D genome organization?
- RNA polymerase II
- CTCF
- DNMT3A
- P53
Correct Answer: CTCF
Q9. Topologically associating domains (TADs) primarily describe which feature?
- Linear gene order conservation only
- Regions of preferential internal chromatin interactions
- Centromere composition
- Mitochondrial genome loops
Correct Answer: Regions of preferential internal chromatin interactions
Q10. Which of the following is a common epigenetic modification associated with gene repression?
- H3K4 trimethylation
- Histone acetylation
- DNA hypomethylation
- H3K27 trimethylation
Correct Answer: H3K27 trimethylation
Q11. Telomeres contain repetitive sequences. What is the human telomeric repeat?
- AAUAAA
- TTAGGG
- CCGCGG
- GGGCCC
Correct Answer: TTAGGG
Q12. Which enzyme maintains telomere length in many cells and is often upregulated in cancer?
- Telomerase
- DNA polymerase delta
- RNA primase
- Topoisomerase II
Correct Answer: Telomerase
Q13. Centromeres are essential for chromosome segregation. They are typically composed of:
- Highly transcribed genes
- Repetitive satellite DNA
- CpG islands only
- Mitochondrial sequences
Correct Answer: Repetitive satellite DNA
Q14. Which genomic elements increase copy number by a “copy-and-paste” mechanism via an RNA intermediate?
- DNA transposons
- Retrotransposons
- Satellite repeats
- Telomeric repeats
Correct Answer: Retrotransposons
Q15. Long non-coding RNAs (lncRNAs) in genome organization can:
- Only code for small peptides
- Assist in chromatin remodeling and gene regulation
- Are exclusively mitochondrial
- Function as replication origins
Correct Answer: Assist in chromatin remodeling and gene regulation
Q16. The C-value paradox refers to:
- The constant GC content across species
- The lack of correlation between genome size and organismal complexity
- The predictable gene number in mammals
- The relationship between chromosome number and body size
Correct Answer: The lack of correlation between genome size and organismal complexity
Q17. Which of the following best describes euchromatic replication timing?
- Late S-phase replication
- Early S-phase replication
- Does not replicate
- Only replicates during G2
Correct Answer: Early S-phase replication
Q18. Which chromatin remodeling complex is known to slide or evict nucleosomes to regulate accessibility?
- RNA-induced silencing complex (RISC)
- SWI/SNF complex
- Ribosome assembly complex
- Telomerase holoenzyme
Correct Answer: SWI/SNF complex
Q19. What is the role of DNA methyltransferase DNMT1 in mammals?
- De novo methylation during development
- Maintenance of methylation during DNA replication
- Removal of methyl groups from DNA
- Histone demethylation
Correct Answer: Maintenance of methylation during DNA replication
Q20. X-chromosome inactivation in female mammals is primarily controlled by:
- Telomerase activity
- XIST lncRNA coating the X chromosome
- Centromere duplication
- MicroRNA degradation
Correct Answer: XIST lncRNA coating the X chromosome
Q21. Which repetitive element class makes up a large fraction of the mammalian genome and can affect gene regulation?
- tRNA genes
- Transposable elements
- Protein-coding exons
- Mitochondrial DNA
Correct Answer: Transposable elements
Q22. A promoter-proximal element that increases transcription regardless of orientation is called a:
- Terminator
- Enhancer
- Origin of replication
- TATA box only
Correct Answer: Enhancer
Q23. Which sequence motif is commonly found in many eukaryotic core promoters and helps recruit transcription machinery?
- Poly-A tail
- TATA box
- Telomeric repeat
- rRNA promoter
Correct Answer: TATA box
Q24. CpG island methylation in gene promoters typically leads to:
- Increased transcription
- Transcriptional silencing
- mRNA splicing changes only
- Telomere elongation
Correct Answer: Transcriptional silencing
Q25. Which feature distinguishes heterochromatin from euchromatin microscopically?
- Heterochromatin stains lightly and is decondensed
- Heterochromatin is highly condensed and stains darkly
- Euchromatin is always repetitive DNA
- Euchromatin is found only in mitochondria
Correct Answer: Heterochromatin is highly condensed and stains darkly
Q26. Which of the following is a histone modification associated with active promoters?
- H3K9 trimethylation
- H3K4 trimethylation
- H3K27 trimethylation
- DNA crosslinking
Correct Answer: H3K4 trimethylation
Q27. Copy number variations (CNVs) in the genome can influence:
- Only mitochondrial function
- Gene dosage and drug response variability
- RNA splicing exclusively
- Only telomere length
Correct Answer: Gene dosage and drug response variability
Q28. In mammals, the majority of gene regulatory elements are located:
- Only within coding exons
- In promoters, enhancers, and distal regulatory regions
- Exclusively in telomeres
- Only in centromeres
Correct Answer: In promoters, enhancers, and distal regulatory regions
Q29. Which molecular technique maps DNA–protein interactions genome-wide and is widely used to study chromatin binding?
- Western blot
- Chromatin immunoprecipitation sequencing (ChIP-seq)
- Mass spectrometry
- Southern blot
Correct Answer: Chromatin immunoprecipitation sequencing (ChIP-seq)
Q30. Mitochondrial genome differs from nuclear genome in that the mitochondrial genome is:
- Linear and larger than nuclear DNA
- Circular and maternally inherited
- Packed into nucleosomes with histones
- Organized into TADs identical to nuclear DNA
Correct Answer: Circular and maternally inherited
Q31. Which histone variant replaces H3 in centromeric nucleosomes and is essential for kinetochore formation?
- H3.3
- CENP-A
- H2AX
- MacroH2A
Correct Answer: CENP-A
Q32. Enhancer RNAs (eRNAs) are thought to:
- Be cleaved into tRNAs
- Play roles in enhancer activity and gene regulation
- Encode ribosomal proteins
- Function as origins of replication
Correct Answer: Play roles in enhancer activity and gene regulation
Q33. Which of the following best explains the concept of gene desert regions?
- Regions with high gene density
- Large genomic regions with few or no genes
- Areas packed with ribosomal genes only
- Telomeric repeats exclusively
Correct Answer: Large genomic regions with few or no genes
Q34. DNAse I hypersensitive sites in chromatin indicate:
- Closed chromatin inaccessible to factors
- Open chromatin accessible to transcription factors
- Regions of telomere repeats
- Sites of DNA replication termination only
Correct Answer: Open chromatin accessible to transcription factors
Q35. Which non-coding RNAs are primarily involved in post-transcriptional gene silencing via RISC?
- rRNAs
- miRNAs
- tRNAs
- snRNAs
Correct Answer: miRNAs
Q36. In pharmacogenomics, variation in which genomic region of CYP genes can alter drug metabolism?
- Only centromeres
- Promoters, exons, introns, and copy number changes
- Telomeres exclusively
- Mitochondrial control region only
Correct Answer: Promoters, exons, introns, and copy number changes
Q37. Which process increases genetic diversity by exchanging DNA between homologous chromosomes during meiosis?
- Transcription
- Homologous recombination (crossing over)
- DNA methylation
- Translation
Correct Answer: Homologous recombination (crossing over)
Q38. Chromosomal translocations often contribute to cancer by:
- Increasing mitochondrial copy number
- Creating oncogenic fusion genes or dysregulating gene expression
- Changing CpG island sequences only
- Inactivating telomerase exclusively
Correct Answer: Creating oncogenic fusion genes or dysregulating gene expression
Q39. Which technique is used to visualize specific DNA sequences on chromosomes in karyotyping?
- FISH (fluorescence in situ hybridization)
- Northern blot
- Chromatin conformation capture
- ELISA
Correct Answer: FISH (fluorescence in situ hybridization)
Q40. Histone H3 variant H3K9me3 is most commonly associated with:
- Active enhancers
- Constitutive heterochromatin and gene silencing
- Spliceosome assembly
- rRNA transcription activation
Correct Answer: Constitutive heterochromatin and gene silencing
Q41. Which of the following describes an orphan receptor-binding enhancer that can act at a distance to regulate gene expression?
- Promoter
- Silencer only
- Distal enhancer
- Origin of replication
Correct Answer: Distal enhancer
Q42. The human genome project revealed approximately how many protein-coding genes in humans?
- ~200,000 genes
- ~20,000 protein-coding genes
- ~1,000 genes
- ~1 million genes
Correct Answer: ~20,000 protein-coding genes
Q43. Which of the following best defines an isoform in the context of gene expression?
- A different species’ genome
- Alternative splice variant of an mRNA producing different protein forms
- DNA replication enzyme variant
- Another name for a promoter
Correct Answer: Alternative splice variant of an mRNA producing different protein forms
Q44. Which genomic feature is most directly studied by Hi-C experiments?
- DNA methylation patterns
- Genome-wide chromatin 3D contacts and interactions
- Protein expression levels
- RNA tertiary structure
Correct Answer: Genome-wide chromatin 3D contacts and interactions
Q45. A promoter that lacks a TATA box often relies on which feature for transcription initiation?
- CpG islands and initiator elements
- Telomere repeats
- tRNA promoters
- Centromeric binding sites
Correct Answer: CpG islands and initiator elements
Q46. Which DNA repair pathway is most important for fixing double-strand breaks using a homologous template?
- Nucleotide excision repair
- Homologous recombination repair
- Base excision repair
- Mismatch repair
Correct Answer: Homologous recombination repair
Q47. The phenomenon where one allele of a gene is expressed preferentially depending on its parental origin is called:
- X-inactivation
- Genomic imprinting
- Alternative splicing
- Epistasis
Correct Answer: Genomic imprinting
Q48. Which of these elements is enriched at nucleolus organizer regions (NORs) and involved in rRNA gene clustering?
- rDNA repeats
- tRNA clusters
- Centromeric satellites
- Enhancer RNAs only
Correct Answer: rDNA repeats
Q49. Which epigenetic therapy targets histone deacetylases to modify chromatin in certain cancers?
- DNMT inhibitors like azacitidine
- HDAC inhibitors like vorinostat
- Telomerase activators
- RNA polymerase inhibitors
Correct Answer: HDAC inhibitors like vorinostat
Q50. Chromatin loops that bring enhancers into proximity with promoters are often stabilized by which combination?
- CTCF and cohesin complexes
- Telomerase and topoisomerase
- Ribosomes and spliceosomes
- DNMTs and HATs only
Correct Answer: CTCF and cohesin complexes

I am a Registered Pharmacist under the Pharmacy Act, 1948, and the founder of PharmacyFreak.com. I hold a Bachelor of Pharmacy degree from Rungta College of Pharmaceutical Science and Research. With a strong academic foundation and practical knowledge, I am committed to providing accurate, easy-to-understand content to support pharmacy students and professionals. My aim is to make complex pharmaceutical concepts accessible and useful for real-world application.
Mail- Sachin@pharmacyfreak.com