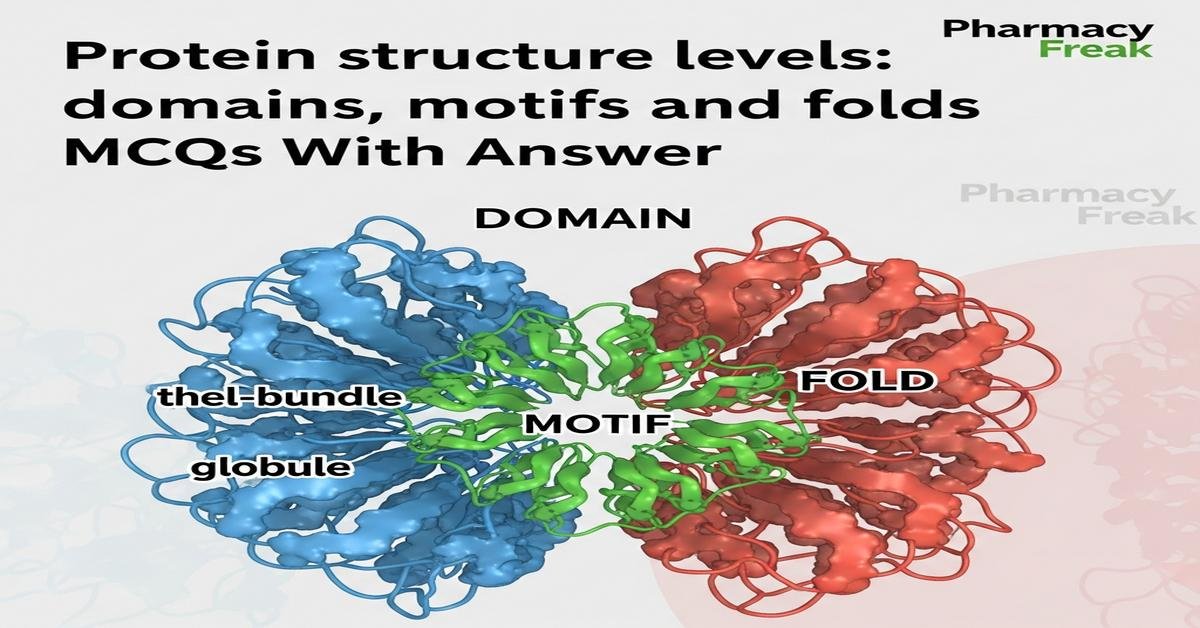
Introduction:
Understanding protein structure at the levels of domains, motifs and folds is essential for modern drug discovery and rational design. Domains are semi-independent structural and functional units; motifs are small, conserved arrangements of secondary structure that often mediate binding or catalysis; folds represent the overall 3D architecture shared by protein families. For M.Pharm students, mastering these concepts helps interpret structure–function relationships, predict drug binding sites, design inhibitors, and anticipate effects of mutations. This set of MCQs focuses on definitions, examples, classification systems (SCOP/CATH), experimental and computational identification, and implications for pharmacology and protein engineering.
Q1. What is the best definition of a protein domain?
- An amino acid sequence motif of 5–10 residues
- A compact, semi-independent folding unit within a protein that often has its own function
- The overall 3D arrangement of secondary structures in a protein family
- A linear segment always located at the N-terminus
Correct Answer: A compact, semi-independent folding unit within a protein that often has its own function
Q2. Which of the following is an example of a common structural motif?
- TIM barrel
- Helix-turn-helix
- Immunoglobulin fold
- Rossmann fold
Correct Answer: Helix-turn-helix
Q3. The TIM (triosephosphate isomerase) barrel is classified as which structural level?
- Primary sequence motif
- Protein fold
- Short linear epitope
- Unstructured region
Correct Answer: Protein fold
Q4. In protein structure databases, which classification focuses primarily on evolutionary relationships and hierarchy of folds?
- BLAST
- SCOP
- Pfam
- BLAT
Correct Answer: SCOP
Q5. Which experimental method provides atomic-resolution data that most directly reveals domains, motifs and folds?
- MALDI-TOF mass spectrometry
- X-ray crystallography
- SDS-PAGE electrophoresis
- UV-visible spectroscopy
Correct Answer: X-ray crystallography
Q6. Which statement most accurately describes a structural motif?
- A fold present only in enzymes
- A short conserved arrangement of secondary structures often associated with a specific function
- A domain boundary defined by proteolytic cleavage
- The entire quaternary assembly of protein subunits
Correct Answer: A short conserved arrangement of secondary structures often associated with a specific function
Q7. The Rossmann fold is commonly associated with binding which ligand type?
- Metal ions like zinc
- Nucleotides such as NAD(P)
- Polysaccharides
- Membrane lipids
Correct Answer: Nucleotides such as NAD(P)
Q8. Pfam is a database primarily used to identify:
- Protein tertiary structures from cryo-EM maps
- Conserved protein domains and families using hidden Markov models
- Post-translational modifications by mass shift
- Small-molecule binding affinities
Correct Answer: Conserved protein domains and families using hidden Markov models
Q9. Which of the following best explains domain shuffling in evolution?
- The irreversible denaturation of domains at high temperature
- The repeated proteolytic cleavage of a single-domain protein
- Loss of secondary structure elements over time
Correct Answer: recombination and rearrangement of existing domains to create new multi-domain proteins
Q10. Which fold is characteristic of antibody variable regions?
- Beta-sandwich immunoglobulin fold
- Alpha-helical bundle
- TIM barrel
- Beta-propeller with seven blades
Correct Answer: Beta-sandwich immunoglobulin fold
Q11. A zinc finger is best classified as which structural feature?
- A protein fold that spans 300 residues
- A small metal-coordinating motif that stabilizes a local structure and mediates DNA binding
- A transmembrane alpha helix
- An intrinsically disordered protein region
Correct Answer: A small metal-coordinating motif that stabilizes a local structure and mediates DNA binding
Q12. How do domains aid rational drug design?
- Domains increase protein solubility but do not influence binding
- Identifying functional domains localizes active or binding sites that can be targeted by drugs
- Domains prevent proteins from interacting with ligands
- Domains only determine mRNA stability, irrelevant to drug binding
Correct Answer: Identifying functional domains localizes active or binding sites that can be targeted by drugs
Q13. Which computational tool or approach is commonly used to predict domain boundaries from sequence?
- Homology modeling of full-length protein without domain annotation
- Domain prediction algorithms combining sequence conservation and predicted secondary structure (e.g., Pfam, CD-Search)
- Mass spectrometry peptide mass fingerprinting
- Chromatography retention time mapping
Correct Answer: Domain prediction algorithms combining sequence conservation and predicted secondary structure (e.g., Pfam, CD-Search)
Q14. In the context of protein folding, what is a folding nucleus?
- The largest domain in a multi-domain protein
- A small set of residues that form early contacts driving the folding pathway toward the native fold
- A motif only present in membrane proteins
- A site where proteases cleave during maturation
Correct Answer: A small set of residues that form early contacts driving the folding pathway toward the native fold
Q15. Which of these structural classification resources organizes proteins by fold topology and evolutionary relationship and complements SCOP?
- PDB (Protein Data Bank)
- CATH
- UniProtKB
- KEGG
Correct Answer: CATH
Q16. Beta-alpha-beta motifs are often indicative of:
- Transmembrane beta-barrels
- Elements of Rossmann-like nucleotide-binding or enzyme active sites
- Intrinsic disorder and flexibility
- Glycosylation sites
Correct Answer: Elements of Rossmann-like nucleotide-binding or enzyme active sites
Q17. Which property most distinguishes a fold from a motif?
- Fold is sequence-based; motif is structure-based
- Fold denotes the global 3D architecture of a protein region; motif is a smaller recurring structural pattern
- Motifs are larger than domains; folds are shorter
- Folds are always enzymatic while motifs are not
Correct Answer: Fold denotes the global 3D architecture of a protein region; motif is a smaller recurring structural pattern
Q18. Domain swapping can lead to which of the following outcomes?
- Formation of oligomers or amyloid-like assemblies with altered function
- Immediate proteasomal degradation of the protein
- Loss of all secondary structure elements permanently
- Conversion of alpha helices into beta strands enzymatically
Correct Answer: Formation of oligomers or amyloid-like assemblies with altered function
Q19. Which statement about short linear motifs (SLiMs) is true?
- SLiMs are large folded domains of >200 residues
- SLiMs are short, often disordered sequence motifs that mediate transient interactions and regulatory events
- SLiMs define the overall fold of an enzyme
- SLiMs always require metal ions to function
Correct Answer: SLiMs are short, often disordered sequence motifs that mediate transient interactions and regulatory events
Q20. Why is knowledge of domain interfaces important when developing protein–protein interaction inhibitors?
- Interfaces are irrelevant because inhibitors only bind active sites
- Domain interfaces contain residues that mediate binding surfaces and are potential sites for small molecules or peptides to disrupt interactions
- Interface residues are always glycosylated and cannot be targeted
- Domain interfaces only form after ligand binding and are therefore unpredictable
Correct Answer: Domain interfaces contain residues that mediate binding surfaces and are potential sites for small molecules or peptides to disrupt interactions

I am a Registered Pharmacist under the Pharmacy Act, 1948, and the founder of PharmacyFreak.com. I hold a Bachelor of Pharmacy degree from Rungta College of Pharmaceutical Science and Research. With a strong academic foundation and practical knowledge, I am committed to providing accurate, easy-to-understand content to support pharmacy students and professionals. My aim is to make complex pharmaceutical concepts accessible and useful for real-world application.
Mail- Sachin@pharmacyfreak.com